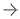
Effect of chronic reductive stress on myocardial proteome. (A) Overall methodology adopted for Tandem Mass Tagged LC-MS/MS analysis. Heart tissues were harvested from non-transgenic (NTg) and caNrf2-Tg (N = 4 mice/group) and protein concentration was determined in homogenates with BCA kit (Bio-Rad, USA). After trypsin digestion, peptides were reconstituted in 0.5 M TEAB and processed for TMT tagging (Tandem Mass Tag kit, ThermoFisher Scientific). LC-MS/MS analysis was performed on a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) equipped with an EASY-Spray nanoESI source. MS and MS/MS data were searched against the amino acid sequence of the Uniprot mouse protein database using Thermo Proteome Discoverer v 2.4.0.305 (Thermo Fisher Scientific). The protein and peptide identification results were further analyzed with Scaffold Q + S v 4.11.1 (Proteome Software Inc.). (B) Comprehensive approach used to calculate the “redox score” in TG by comparing to basal redox factors in NTg. Here, we included the levels of small molecular antioxidants (i.e. GSH, Cysteine/Cystine ratio); antioxidant proteins (i.e. GST MU, NQO1, CAT, GPX 1, SOD1, GCLM, GCLC, SOD2); antioxidant enzymatic levels (i.e. TAC); antioxidant gene expression (i.e. GCLM, NQO1, GSR, GST alpha, GCLC, GCLM, GSR, NQO1, GST MU, GPX 1, CAT) as well as the reactive oxygen species levels (i.e. DHE). (C) Quantitative genotyping in caNrf2 mice caNrf2 genotyping primer, endogenous primers for Nrf2, NQO1 and GCLC. (D) Principal Component Analysis (PCA) Plot generated using Total Identified Proteins (TIPs) showed segregation of NTg and CaNrf2 TG as distinct groups. (E) Venn diagram showing the number of Differentially Expressed Proteins (DEPs) based on different fold change (CaNrf2 TG vs. NTg) identified in TMT proteome software. (F) Global heat map generated using R studio for TIPs (1,105) proteins identified in TMT analysis. (G) Gene ontology pathway by PANTHER analysis. Protein Analysis THrough Evolutionary Relationships (PANTHER) analysis (http://pantherdb.org/) for biological function distributed the total identified proteins (TIPs) into different metabolic categories. About 25% of the proteome function grouped under metabolite interconversion category, with oxidoreductase enzyme family as the top upregulated one. The proteins identified under oxidoreductase group are shown in (H).
August 8, 2022 — Two years ago, University of Alabama at Birmingham researchers and colleagues reported that reductive stress — an imbalance in the normal oxidation/reduction, or redox, homeostasis — caused pathological changes associated with heart failure in a mouse model. This was a follow-up to their 2018 clinical study that found about one in six heart failure patients shows reductive stress.
Now they have extended their description of changes caused by reductive stress to describe changes in the proteome of heart cells in mice, disclosing a likely proteome signature for reductive stress cardiomyopathy. A proteome is the complement of proteins expressed in a cell or tissue.
Using tandem mass spectrometry, researchers led by Rajasekaran Namakkal-Soorappan, Ph.D., associate professor in the UAB Department of Pathology, Division of Molecular and Cellular Pathology, looked at differential protein expression between control hearts and reductive-stress hearts in a mouse model of chronic reductive stress.
They found about 560 proteins were differentially expressed, and 32 proteins were significantly altered — 20 being upregulated and 12 downregulated. The reductive stress mouse model is caused by a constitutively active NRF2, the redox sensor that maintains redox homeostasis in cells.
Through gene ontology and pathway analysis, the researchers found that the majority of the differentially expressed proteins are involved in stress-related pathways such as antioxidants, NADPH, protein quality control and others. Proteins involved in mitochondrial respiration, lipophagy and cardiac rhythm were dramatically decreased in the reductive stress hearts.
The most significantly changed subset of proteins was in the glutathione family. Glutathione is an antioxidant, active in redox homeostasis, that can exist in a reduced or oxidized form.
Surprisingly, the levels of about half of 104 altered proteins were found not to correlate with levels of their messenger RNAs, the gene message that is read by ribosomes to make a protein. The reason for this asynchrony is not known.
In association with the altered proteome, the reductive stress mice displayed pathological cardiac remodeling. This cardiomyopathy makes it harder for the heart to pump blood, and it can lead to heart failure. The researchers also found post-translational modifications such as oxidation, N-ethylmaleimide, methionine loss and acetylation in the reductive stress hearts.
“Under reductive stress, we observed downregulation of several myocardial adaptation or rescue pathways and upregulation of pathophysiological processes, which are associated with reductive stress cardiomyopathy over time,” Namakkal-Soorappan said. “Thus, our results provide a rationale to develop personalized antioxidant therapeutic strategies to avoid reductive stress-mediated proteome alterations in humans.”
The report, “Tandem Mass Tagging based identification of proteome signatures for reductive stress cardiomyopathy,” is published in Frontiers in Cardiovascular Medicine.
For more information: https://www.uab.edu/


 February 03, 2026
February 03, 2026